CRISPR-Cas9 has emerged as a potent genome editing tool that promises unprecedented control and specificity in targeting desired sequences and has been successfully employed in many cell lines and organisms. However, whether CRISPR-Cas9 can function in a setting where the target sequences can fold into stable secondary structures (alternative structures to the well-known double-helix DNA) is not known. In this project, researchers from Physics and Chemistry departments will establish capabilities and limitations of CRISPR-Cas9 in targeting such sequences and functioning in the vicinity of alternative secondary structures.
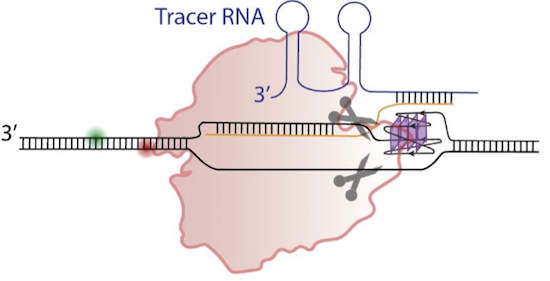
Drs. Balci and Basu have received a three-year grant from the National Institutes of Health (NIH) of approximately $454k to perform their research. Using single molecule and ensemble level methods, the Balci and Basu Labs will perform in vitro and in cellulo experiments to establish the capabilities and limitations of nuclease-dead mutant of Cas9 (dCas9) to target G-quadruplex forming sequences in the promoter regions and regulate transcription. Tyrosine hydroxylase (the rate limiting enzyme in dopamine biosynthesis), c-MYC (a proto-oncogene that codes for a transcription factor), and KRAS (a significant gene in signaling and cell proliferation) will be the test systems for this new technology.

